Research Overview
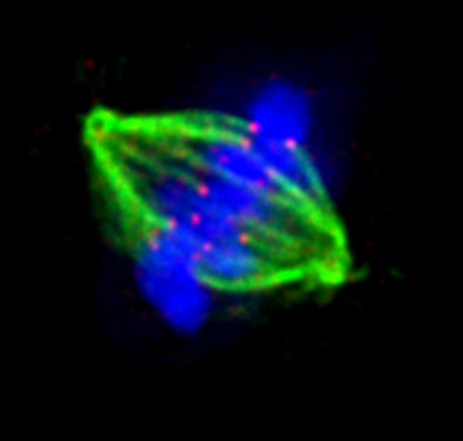
Regulated sub-cellular movements are a fundamental aspect of all living cells and they rely on precisely controlled force generation mechanisms. How are forces imparted, monitored and corrected in response to heterogeneous biochemical and physical cues is a fascinating and challenging biological problem. We address these fundamental questions in the context of dividing human cells and apply the molecular knowledge on force generation and cell division to accelerate cytoskeletal drug discovery and drug resistance biomarker research. Defective force generation during cell division can lead to chromosomal instability and errors in the size, content and fate of daughter cells. Our research findings are therefore relevant to the understanding of irregular chromosome numbers (aneuploidy) and tissue disorganisation found in aggressive cancers and several age-related disorders.
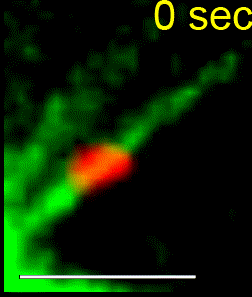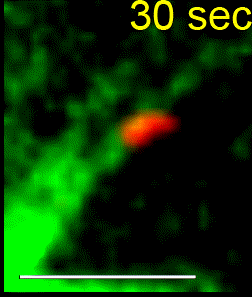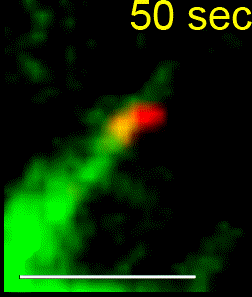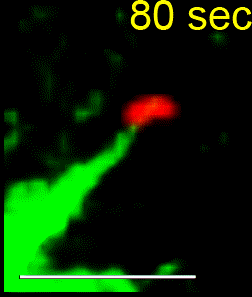
We combine single-cell microscopy with molecular and biochemical approaches. We collaboratively develop computational tools to extract single-cell and population metrics. These approaches form the platform for two streams of studies in our group: (i) To investigate how microtubules are correctly anchored, and subsequently how force generation powers the movements of chromosomes and the mitotic spindle, we use high and super-resolution live-cell imaging of human cells. (ii) To translate the basic knowledge on mitotic microtubule regulation into accelerating the discovery of novel microtubule perturbing drugs and drug resistance biomarkers, we have adopted bioinformatics approaches. For example, pharmacogenomics studies have allowed us to build transcriptional signatures to predict cells sensitive to Paclitaxel and to reposition two FDA-approved drugs as microtubule stabilisers (Splitomycin and Glipizide) through drug repositioning (Iorio et al., 2015). Bioinformatic analysis of human genetic variants of kinetochore and microtubule-associated proteins has led us to uncover protein residues or regions crucial for chromosomal and genomic stability, taking us a step closer to precision medicine (see CIVa database of variants screened from >150,000 healthy individuals and patients).

Another wider aim of our research is to better understand how cells successfully position their spindle to ensure proper cell division in order to generate two genetically identical daughter cells from a single mother cell. Mispositioning of the mitotic spindle can lead to an incorrect plane of cell division and consequently altered stem cell fate and disorganized epithelia. In knockdown studies, our lab uncovered a novel role for MARK2 in maintaining the spindle at the cell’s geometric center. Following MARK2 depletion, spindles glide along the cell cortex, leading to a failure in identifying the correct division plane. For more information please visit our recent Publication List.
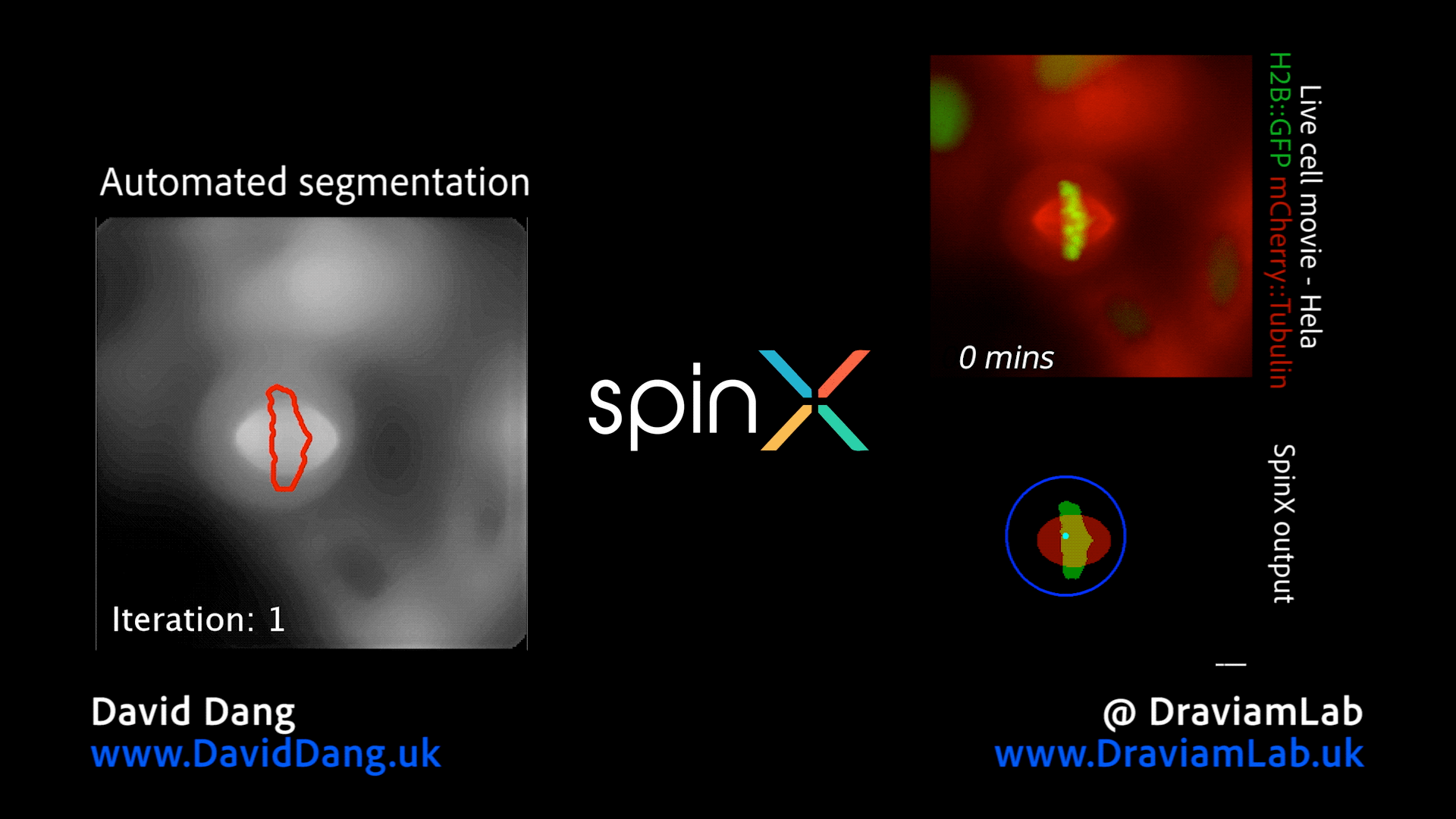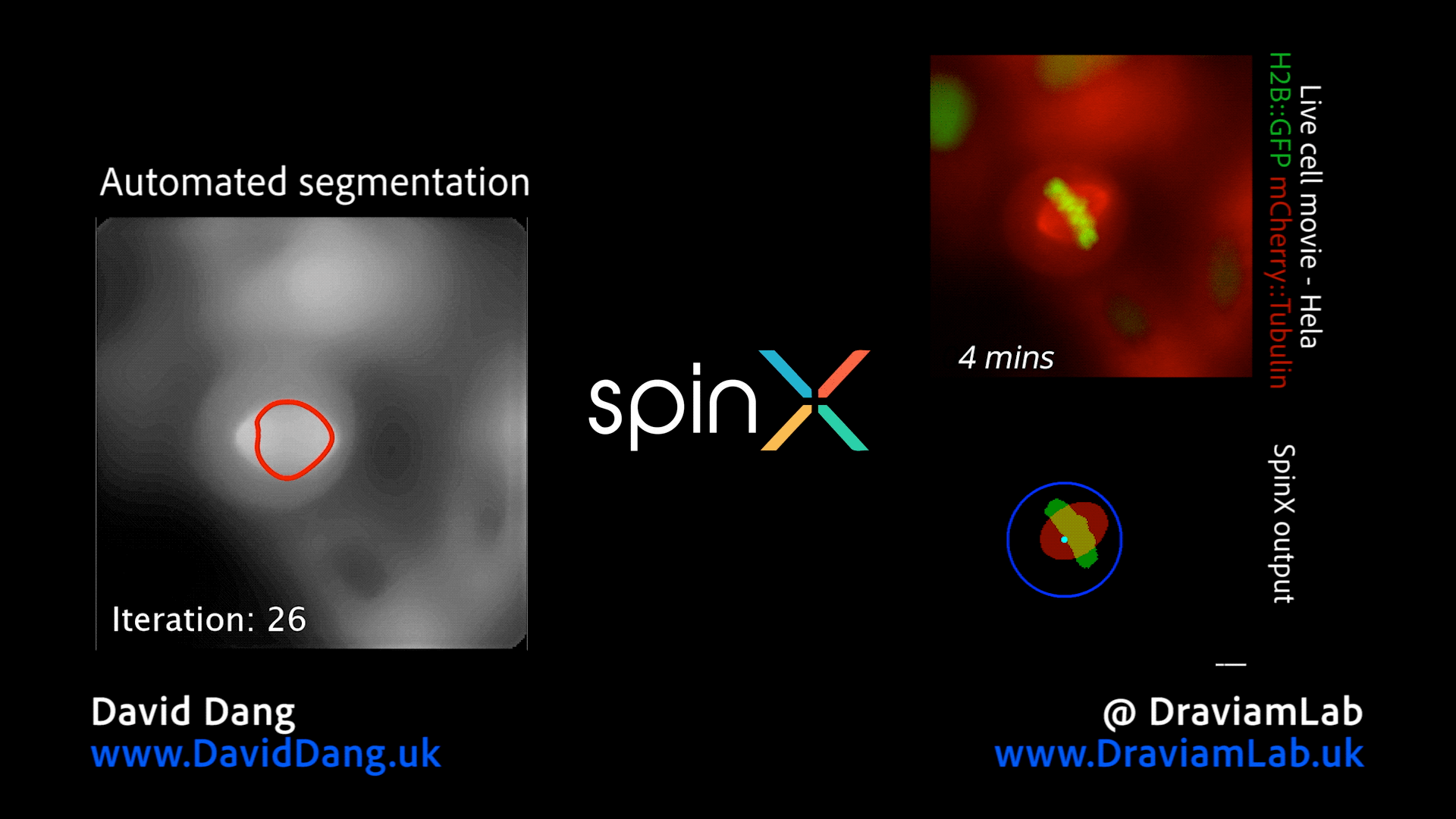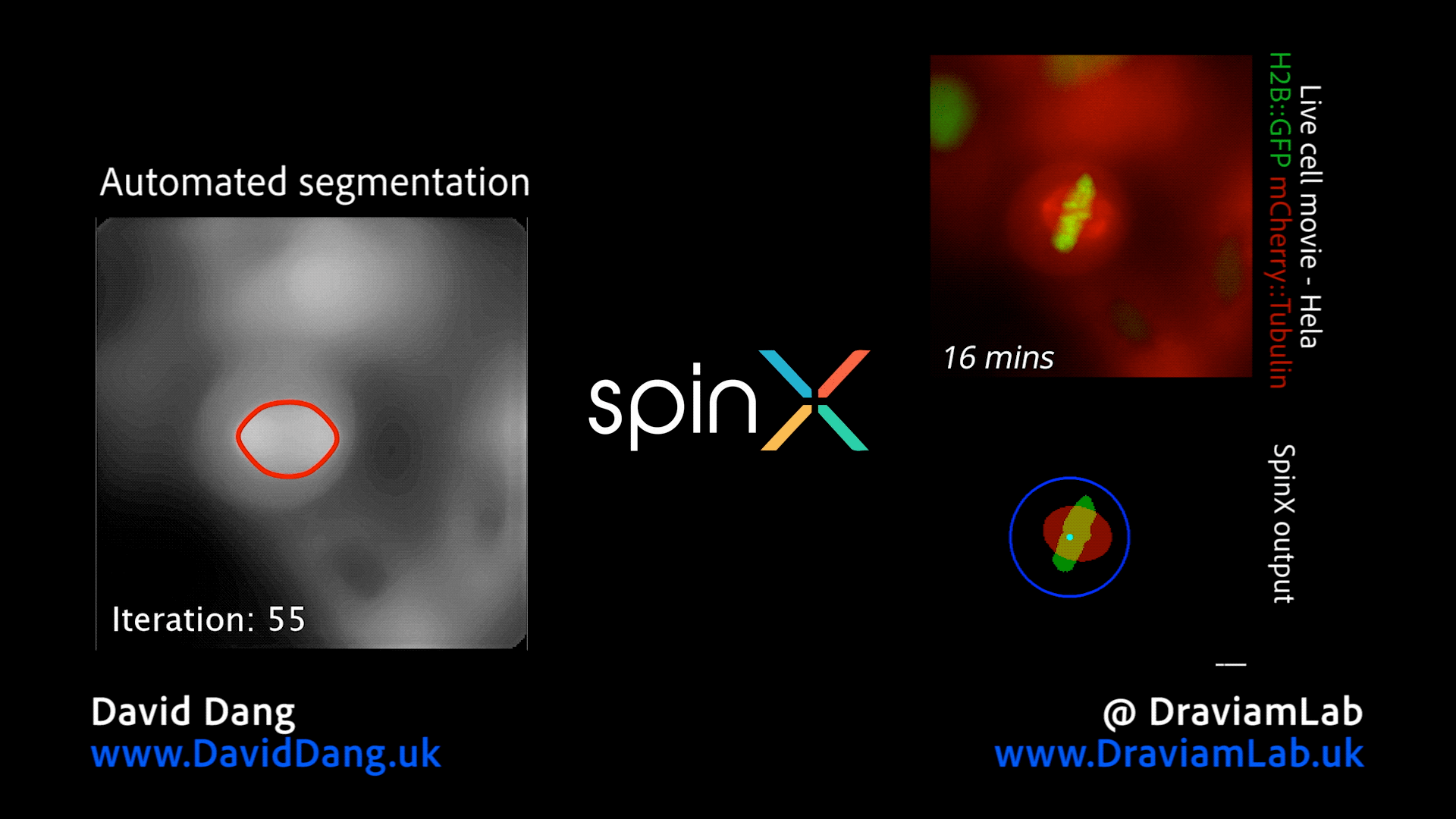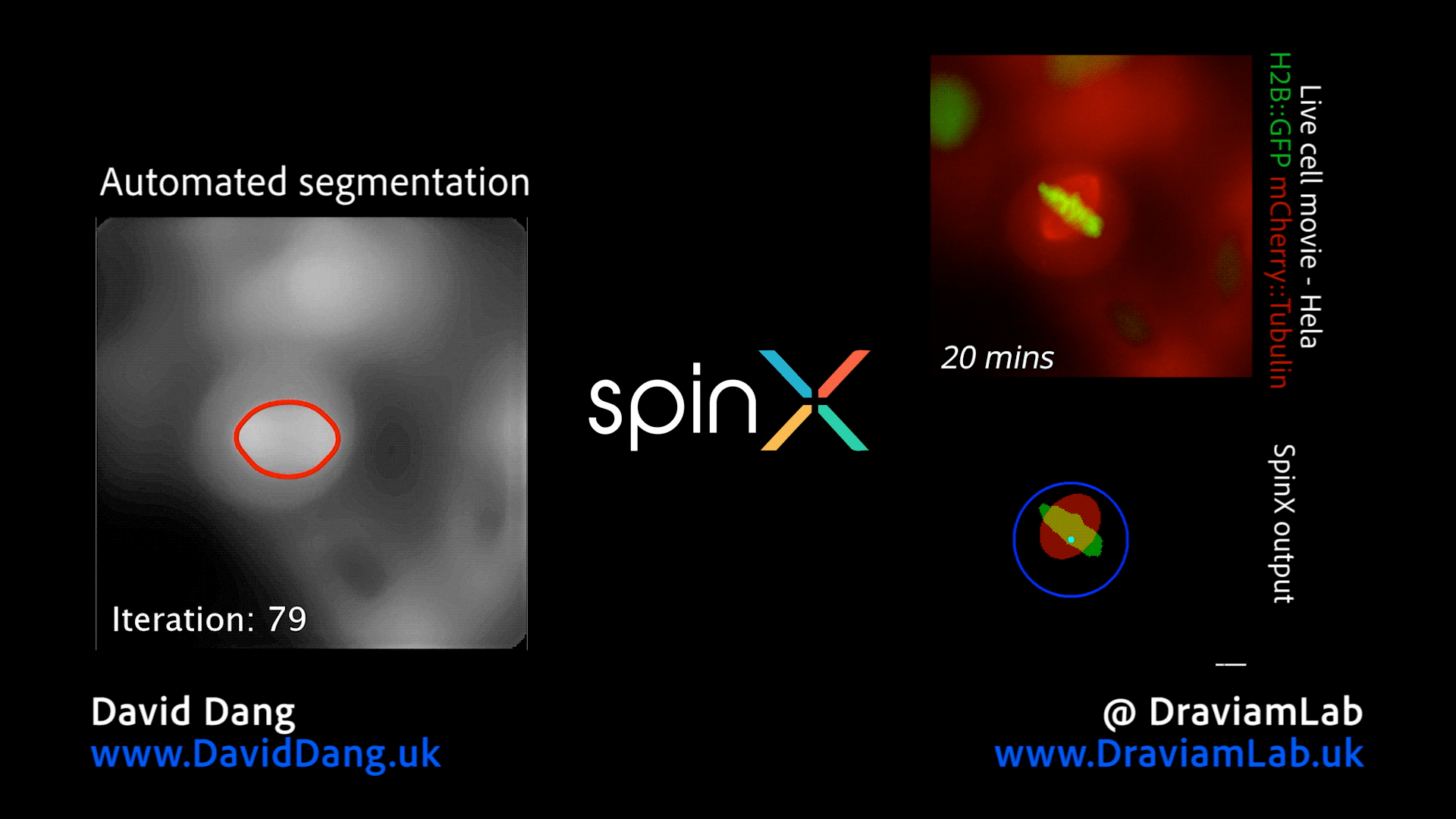
By augmenting conventional image segmentation protocols with Deep Learning methods, we have developed the SpinX software to precisely track spindle movement through time. This computational approach allows us to monitor changes in spindle movements in response to drug treatments or protein depletions. The software is available on APEER and is being generalised further with ZeissTM UK and Germany. The software will help automate the analysis of spindle dynamics in CRISPR-edited iPSC lines expressing human genome variants of cell cycle regulators.